What haplotypes and assembly patches can I see for human?
Where do haplotypes and patches come from?
The GRC provides haplotypic regions along with the Primary assembly for human. In addition to haplotypes, which are regions with known variations to the Primary assembly, the GRC provides assembly patches on a regular basis.
Patches and Haplotypes in the Human Genome
There are two types of assembly patches: Novel patches and Fix patches. Novel patches are additional sequence for alternate alleles that are not represented on the primary assembly. Fix patches are additional sequence that will replace the known regions of misassembly in GRCh38 when the next major assembly update is released.
How is this shown on the browser?
You might notice some strange-looking chromosome names, or regions of the genome that have red or green highlighting. Red regions represent regions of the genome where there is a haplotype or novel patch. Green regions represent regions of the genome where there is a fix patch. For an example, see the top image in Region in Detail. You can jump to haplotypes and patches by clicking on those highlighted regions. Other access points include BioMart (see the Region: Chromosome filter), and the Perl API.
What haplotypes are available with GRCh38?
A list of haplotypes and patches can be found on the GRC human overview page. There are over 200 alternate haplotypes available for human in GRCh38.
How can I download the DNA sequence for haplotypes and patches?
The DNA sequence for the primary assembly plus haplotypes and patches can be downloaded from our FTP site.
In addition to the primary assembly chromosomes, we have constructed 'patched chromosomes' by applying the individual haplotypes and patches to their relevant chromosome at the position indicated by the GRC as being the 'equivalent' region. Each patched chromosome has only one patch or one haplotype applied. The patched chromosome will have a length that is similar to the primary assembly chromosome; if the length of the assembly patch is shorter than the region it replaces, then the patch chromosome will also be shorter than the primary assembly chromosome.
Outside the region of the assembly patch, the entire length of a 'patch chromosome' is padded with Ns. All 'patch chromosomes' in Ensembl have their sequence padded with N's to ensure alignment programs can report the correct index regions e.g. A patch with a start position of 1,000,001 will have 1e6 N's added its start so an alignment program will report coordinates with respect to the whole chromosome.
How do you (Ensembl) know where to apply an assembly patch?
When the GRC submit the assembly patches, they specify the genomic location (ie. chromosome, plus start- and end-coordinates) on the primary assembly that the assembly patch relates to. We download this information from the GRC FTP site.
How do I know which parts of the patch are different compared to the chromosome on the primary assembly?
If you are interested in alignments, we show these in two ways:
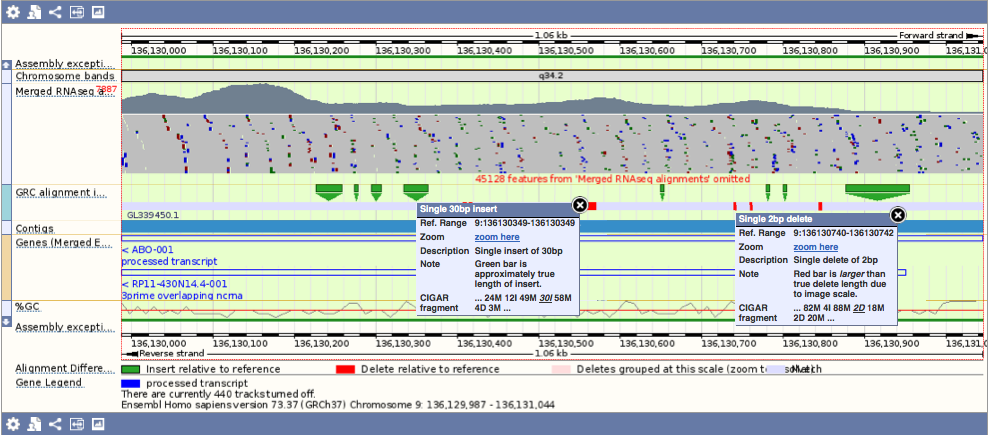
We download GRC's alignments between the primary assembly chromosome and assembly patch, and display these alignments on Region In Detail in the "GRC alignment import" track. This track can only be seen when you're viewing a primary assembly chromosome in a region where there is an overlapping assembly patch. Here is example for the ABO gene. You can click on the red lines and green triangles for more information.
We also produce our own alignments between the primary assembly chromosome and assembly patch, using LASTZ. You can see these alignments as a pink track when you're comparing the primary assembly and the patch in 'Region Comparison' view.
How do I access the haplotypes and patches programmatically?
The haplotypes and assembly patches can be fetch using our API. For historical reasons, when using the API the primary assembly is known as the ‘reference’ sequence and the alternate sequences (haplotypes and patches) are know as ‘non-reference’ sequence. For example:
$slices = $slice_adaptor->fetch_all( ‘toplevel’, undef, 1 );
or
$assembly_exception_features = $assembly_exception_feature_adaptor->fetch_all_by_Slice($slice);
How do I access the haplotypes and patches using MySQL queries?
We store information about alternate sequences in the assembly_exception table.
mysql -uanonymous -hensembldb.ensembl.org -P3306 -Dhomo_sapiens_core_77_38 -e "select sr2.name as chr_name, exc_seq_region_start,exc_seq_region_end,exc_type,sr1.name as alternate_seq_name,seq_region_start, seq_region_end from assembly_exception ae, seq_region sr1, seq_region sr2 where sr1.seq_region_id=ae.seq_region_id and sr2.seq_region_id=ae.exc_seq_region_id order by chr_name,exc_seq_region_start"
For more about patches and haplotypes, see our blog post.
If you have any other questions about Ensembl, please do not hesitate to contact our HelpDesk. You may also like to subscribe to the developers' mailing list.



