How do I access RefSeq annotation in Ensembl?
Ensembl gene sets are comprehensive sets, based on supporting evidence from sequence databases including UniProt and RefSeq. Where a transcript in the Ensembl set has a close match to a RefSeq transcript, the two transcripts are linked. RefSeq IDs linked to Ensembl transcripts are available in the browser under the Transcript tab, General identifiers view, and also from BioMart and from the API as Xrefs. Nearly 100% of NCBI RefSeq proteins have a corresponding protein in the Ensembl annotation.
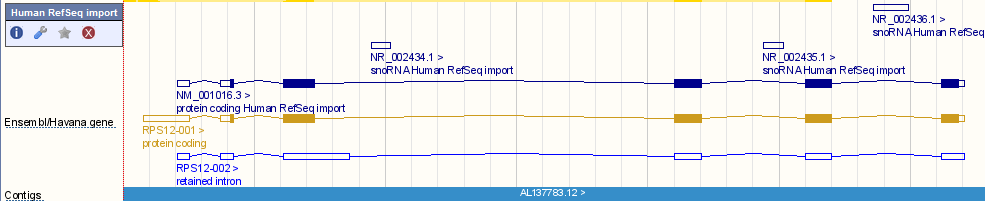
In addition to linking the Ensembl annotation to the corresponding RefSeq annotation, the complete set of RefSeq models are imported into Ensembl for human and mouse. These are visible as a separate track in Location tab. To switch on the track, click 'Configure this page, open the 'Genes' list, and select 'Human RefSeq import' or 'Mouse RefSeq import'. The image below shows there are imported RefSeq models reflecting one protein coding transcript and three noncoding RNAs (snoRNAs). The Ensembl/Havana gene track shows one protein coding transcript agreed on by both the Ensembl annotation pipeline and Havana manual curation. A second transcript has a retained intron and is untranslated.
We load these models directly into the otherfeatures database and do not change any coordinates. Click here for an example script to access RefSeq gene models for human using the API.
Why do they differ?
While Ensembl gene models are annotated directly on the reference genome, RefSeq annotates on mRNA sequences. Due to sequence differences between the reference genomes and individual mRNAs, some of the RefSeq mRNAs may not map perfectly to the reference genome. For example, translations may contain stop codons when they are translated from the reference genome's DNA. Ensembl transcripts will reflect the reference genome in these cases, not the mRNA, and therefore there can be small differences between RefSeq mRNA/proteins and Ensembl transcripts/proteins.
See this article for more on Ensembl gene annotation.
If you have any other questions about Ensembl, please do not hesitate to contact our HelpDesk. You may also like to subscribe to the developers' mailing list.


