
Human
Homo sapiens
Annotation progress
Click on a chromosome to browse:
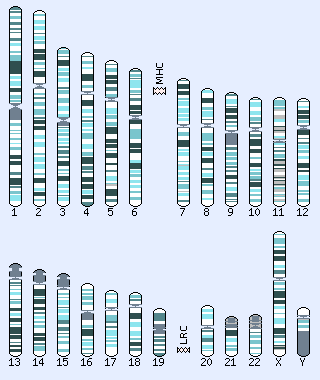
Shading indicates the karyotype bands
This Release
| Release Date | 7 February 2017 |
| Datafreeze Date | 18 October 2016 |
| As used in | GENCODE 26; Ensembl 88 genebuild |
| Reference Assembly | GRCh38 |
| Annotation Method | Complete first pass Manual Annotation |
Comparative Analysis
More info...
| Region | Available species / haplotypes / strains | Examples |
|---|---|---|
| MHC | rat, dog, chimpanzee, gorilla, human (ref. plus 7 haplotypes), mouse (ref. plus 2 NOD strains), pig (Duroc & Large White), rat (mixed sex strain), tasmanian devil, wallaby | example |
| LRC | pig, gorilla, human (ref. and 8 haplotypes). | example |
| Autosome | human chr 1 and mouse chr 4; human chr 17 and mouse chr 11 | 1/4 17/11 |
| Allosome | X: human, mouse and pig | example |
| human chr 20, mouse chr 2 and approx 10 Mbp of pig chr 17 | example |
Loss of Function Variants
884 transcripts with SNVs, identified in human genomes as part of the pilot phase of the 1000 genome project, were manually annotated for their predicted functional effects. These are shown in a separate track on Vega, and the names of the genes / transcripts are prefixed with 'LOF:'. Further information.

 Example transcript
Example transcript
 Example region
Example region
 Example gene
Example gene
